Machine learning is usually applied to man-made targets — masses of data about the contents of a digital image, or the movement of cars through a city. This week, however, a new study applied the approach to find order in an equally chaotic mass of data: the cell. By using an advanced machine learning approach, a team of researchers from the University of Texas was able to sift through years of experimental data to understand how tadpole cells control their own development, and then used that understanding to take control of the cells themselves. It’s a breakthrough they think they can apply directly to developing all new treatments for cancer.
When it comes to cancer, cellular control is the whole ball game. A normal cell responds to signals from the body and plays by a certain set of rules that governs how it functions and reproduces. A cancer cell, on the other hand, refuses to play by the rules, reproducing recklessly, pursuing short-term evolutionary goals that hurt the organism overall. But what if scientists could simply change the cell’s rules back to normal, or prescribe drugs that would specifically combat the cancer’s new rules? The problem is, as easy as it is to observe the change in the cell’s behavior, it’s very difficult for scientists to figure out the particular interconnected sequence of actual molecules make those behaviors happen. That’s where machine learning comes in.
The team created a neural network infrastructure where each neuron was a chemical interaction in tadpole cells, then trained the neural network by having it check its own outcomes against the “truth” answers provided by prior experiments.
Each individual prior experiment only revealed the role of a particular signal molecule or receptor, like tiny pieces to a puzzle. But over time, this neural network was able to find associations between the bits of data, and then decipher where they fit into the larger puzzle of biochemical pathways and cellular outcomes.
The A.I.'s inferred control mechanism. It was able to sniff out not only associations between nodes, but self-control feedback loops, as well.
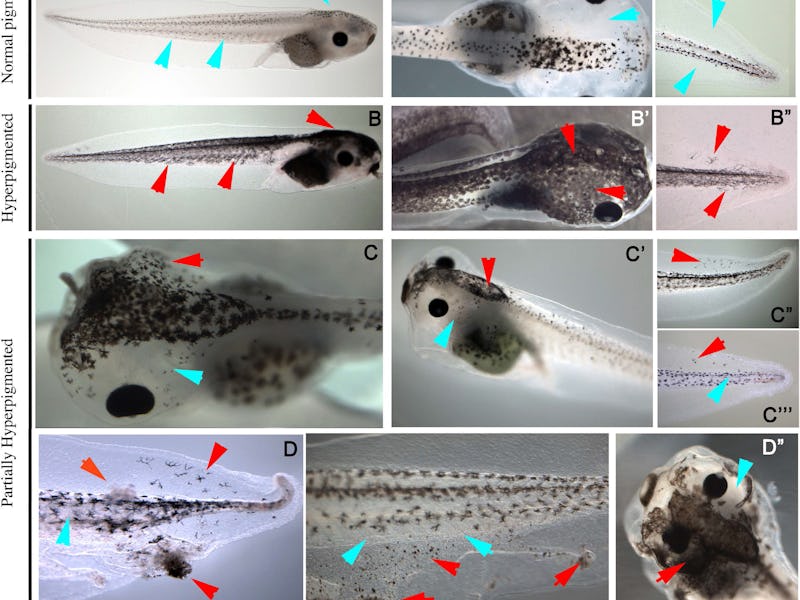
In this study, the proof of concept was using their A.I.’s new insights to design a drug that could specifically turn some tadpole cells a telltale “melanoma-like” color, and others not. Administering this drug traditionally results in an all-or-nothing color change, every cell or no cells in a particular tadpole; by predicting not only the spotted pattern but also the percentage of the tadpole cells that would change color, the team has provided strong evidence that their model has real predictive validity.
So, now, the goal will be to build databases of properly annotated information about control points on certain cancer cell types, and start to train deep learning neural networks in how those points all work together. Happily, cancer research is one of the most data-saturated areas of study in the world today, so the backlog of experimental data should be easy enough to find.
For now, this A.I. has only studied and affected tadpole cells, but notice that it was then used to design a drug that could produce a cancer-related change. When applied to a human cell type and told to understand its control, a similar application could allow design of an all-new cancer medicine for testing.
That’s the sort of thing that could really let lifespans extend by decades: not newly advanced drugs, but newly advanced tools for drug development. It’s a major area of research, with investment from universities and giants like Microsoft, alike. There will never be any one single cure for all cancers — but it is possible there could someday be one single A.I. “researcher” that can figure them all out.